
Cell biology of disease. Muscle physiology and pathology.
Keywords: Membrane dynamics, RNA metabolism, neuromuscular diseases
Our main research interest is to understand how RNA-related proteins impact cellular/tissue homeostasis in health and disease.
Ongoing studies aim to assess the intriguing interplay between RNA metabolism and membrane trafficking regulating gene expression at subcellular level. We use a multi-disciplinary approach involving cell imaging and biochemical methods to isolate and characterize RNA-protein complexes associated to different organelles and membrane subdomains. We have a long-standing interest in neuromuscular pathologies, such as Duchenne Muscular Dystrophy (DMD) and Spinal Muscular Atrophy (SMA), to provide new insight into pathogenic mechanisms as well as to develop therapeutic strategies
Education:
1994: Degree in Biology, Sapienza University of Rome (110/110 cum laude).
1999: Master of Medicine (Clinical Pathology), Faculty of Medicine, Sapienza University of Rome (70/70 cum laude).
2008: PhD, Biotechnology and Biomedicine, University of L’Aquila, Department of Medicine.
Positions:
1992-1994 Research training at Sapienza University of Rome, Department ofExperimental Medicine.
1995-2000: Research fellow at National Research Council-ITBM, Rome.
2001-2005: Research fellow at National Research Council-INMM, Rome.
2005-2010: Post-doctoral fellow at National Research Council-IBPM, Rome.
2010 up to now: Researcher (permanent position) at National Research Council, Rome. Research Project P.I. Neuromuscular Diseases. Translational control and membrane remodelling: role of RNA-binding proteins.
Honors:
2009 Award for Best Poster at Telethon Conventions, Italy.
2011 Award for Best Poster at Telethon Conventions, Italy.
2013 Award for Best Poster at Telethon Conventions, Italy.
Patents:
The following patents, all active, are related to Duchenne Muscular Dystrophy. Collaboration agreement between CNR and Zingenix Ltd (prot CNR-IBPM n. 0000912, 16/04/2019).
- Granted European Patent n. EP3194600B1 (2019). Granted USA patent n. US10301367B2 (2019). Title. “Compositions and Methods for treatment of Muscular Dystrophy”. International publication number: WO2016/016119.
- Granted European patent n. EP3030666B1 (2019). Granted USA patent n. US10286085B2 (2019).
Title: “Compositions and Methods for treatment of Muscular Dystrophy”. International publication number: WO2015/018503. Originated from Italian patent application n TO2013A000669, 2013.
- Granted U.S. patent n. 304.235 (2012). Granted European patent n. EP2193200B1 (2011). Granted Italian patent n. 1379230.
Title: Nucleic acid codifying for a utrophin transcription specific regulating protein, protein codified thereby and uses thereof. Validated in Italy, Germany, France and UK (from PCT/IB2008/054089 filed on 2008).
- Gabanella F, Colizza A, Mottola MC, Francati S, Blaconà G, Petrella C, Barbato C, Greco A, Ralli M, Fiore M, Corbi N, Ferraguti G, Corsi A, Minni A, de Vincentiis M, Passananti C, Di Certo MG. The RNA-Binding Protein SMN as a Novel Player in Laryngeal Squamous Cell Carcinoma. Int J Mol Sci. 2023 Jan 16;24(2):1794. doi: 10.3390/ijms24021794.
- Gabanella F, Barbato C, Corbi N, Fiore M, Petrella C, de Vincentiis M, Greco A, Ferraguti G, Corsi A, Ralli M, Pecorella I, Di Gioia C, Pecorini F, Brunelli R, Passananti C, Minni A, Di Certo MG. Exploring Mitochondrial Localization of SARS-CoV-2 RNA by Padlock Assay: A Pilot Study in Human Placenta. Int J Mol Sci. 2022 Feb 14;23(4):2100. doi: 10.3390/ijms23042100.
- Gabanella F, Barbato C, Fiore M, Petrella C, de Vincentiis M, Greco A, Minni A, Corbi N, Passananti C, Di Certo MG. Fine-Tuning of mTOR mRNA and Nucleolin Complexes by SMN. 2021 Nov 4;10(11):3015. doi: 10.3390/cells10113015.
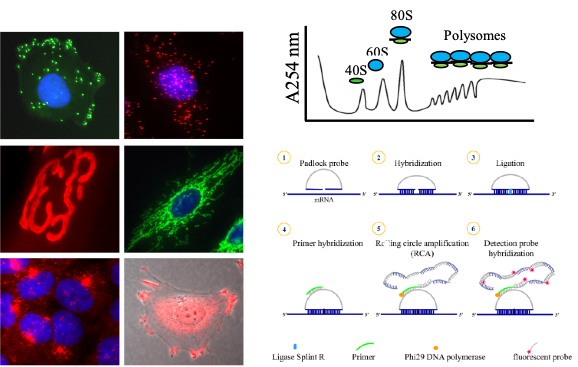
- Gabanella F, Onori A, Ralli M, Greco A, Passananti C, Di Certo MG. SMN protein promotes membrane compartmentalization of ribosomal protein S6 transcript in human fibroblasts. Sci Rep. 2020 Nov 4;10(1):19000. doi: 10.1038/s41598-020-76174-3.
- Gabanella F, Pisani C, Borreca A, Farioli-Vecchioli S, Ciotti MT, Ingegnere T, Onori A, Ammassari-Teule M, Corbi N, Canu N, Monaco L, Passananti C, Di Certo MG. SMN affects membrane remodelling and anchoring of the protein synthesis machinery. J Cell Sci. 2016 Feb 15;129(4):804-16. doi: 10.1242/jcs.176750.

2018-2021 Lazio Innova Project 15286 Passananti (PI). Title: “Geni artificiali come strategia terapeutica per la Distrofia muscolare di Duchenne”. (Artificial genes as therapeutic strategy for Duchenne Muscular Dystrophy). (Role: Co-Investigator).
2017-2018 InterOmics Project 19699 Passananti (PI). Project title: “Cell-based Omics approaches to identify pathways implicated in Duchenne Muscular Dystrophy by the use of therapeutic artificial genes”. (Role: Co-Investigator).
2017-2019 Scientific Cooperation Agreement CNR-CONICET 11985 Corbi (PI). Project title: “Mechanisms of transcription in memory persistence. Role of NF-kB and Che-1” (Role: Co-Investigator).
2014-2017 Telethon project GGP14073 Corbi (PI). Project title: “Innovative therapeutic strategy for Duchenne Muscular Dystrophy by AAV mediated delivery of artificial transcription factor genes.” (Role: Co-Investigator).
2013-2015 Association Française contre les Myopathies (AFM) grant number 15586 Monaco (PI). Project title: “PDE inhibitors as possible pharmacological therapy for muscular dystrophy.” (Role: Co-Investigator).
2011/2012 Telethon Project GGP10094 Mattei (PI). Project title: “Experimental gene therapy of Duchenne Muscular Dystrophy by artificial transcription factors upregulating the dystrophin-related Utrophin.” (Role: Co-Investigator).
2010/2013 AIRC Project IG2009 Passananti (PI). Project title: “Anti-apoptotic role of Che-1/Hax1 interaction in sever congenital neutropenia (SCN), a preleukemic condition.” (Role: Co-Investigator).
2007-2009 Telethon project GGP07177 Mattei (PI). Project title: “Animal models to validate the use of new small synthetic transcription factors in the gene therapy of muscle dystrophies.” (Role: Co-Investigator).
