Group Leader : Emilia Vitale
Topics: Neurobiology
The NeurOMICS lab is dedicated to advancing the understanding of the complexities of dementia and to generating new biological insights into the development of personalized therapeutic strategies for neurodegenerative diseases and dementia.
Dementia is an enormous burden for patients, patients’ families and the health-care system. Dementia is not a single disease but rather, it represents a descriptive term for a collection of symptoms that can be caused by a number of disorders that affect the brain. People with dementia have significantly impaired intellectual functioning that interferes with normal activities and relationships. While memory loss is a common symptom of dementia, by itself it is not sufficient to assign a diagnosis of dementia to a person. The diagnosis is based on impairment of memory and language skills while consciousness remains maintained. Changes in the brains of people with these disorders may begin many years before memory loss or the appearance of other symptoms.
BioMarkers. Many types of biomarker tests have been used in aging research and related dementias to detect these brain changes in people. Finding these changes early in the disease process may help identify people who are at the greatest risk of Alzheimer or other dementias and may help identify those who may benefit most from a particular treatment. Our major interest is to identify genomic variants associated with disease and analyze the effects of mutations that cause different forms of Dementia.
Goals. Our goal is to apply this knowledge to the analysis of those genes in which we have strong candidate causative variants and the corresponding affected pathways. Together, these data will also provide us with a training set for identifying classification signatures specific to each pathway, which we will then use to study the roles and mechanisms of pathogenicity of genes not previously associated with Dementia.
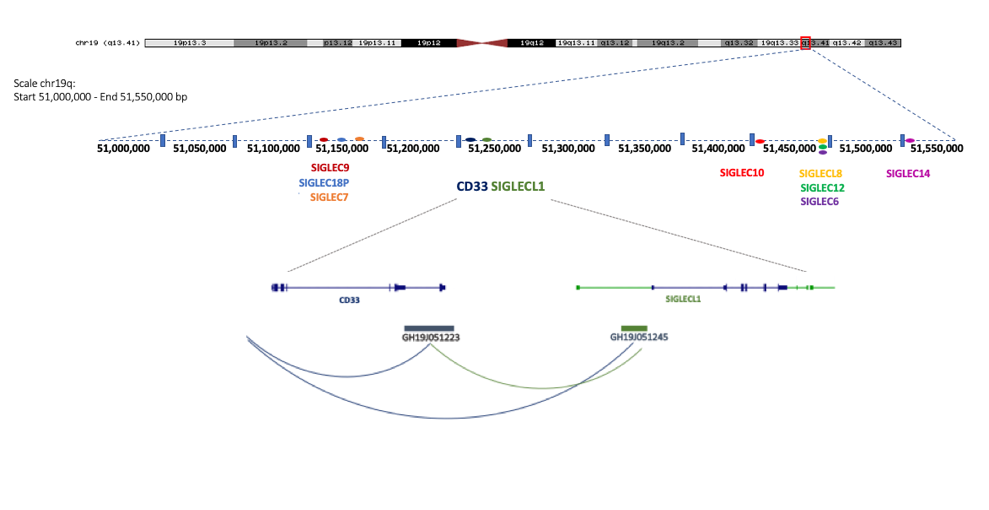
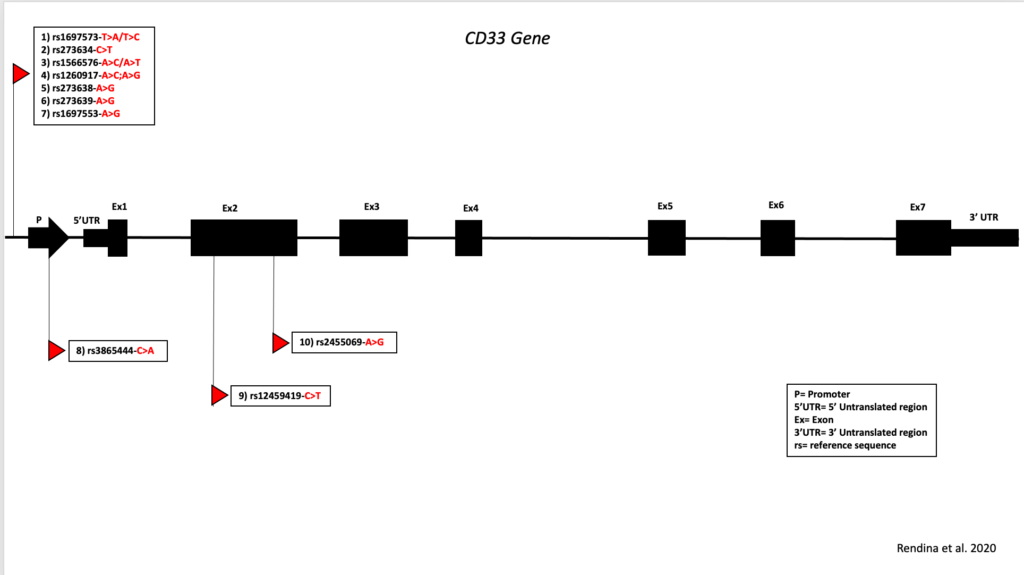
iPSCs. Mutations in genes related to neuroinflammation may be predisposing factors in these diseases. Identification of the coding and non-coding transcriptional neuronal signatures in dementia associated with haploinsufficiency in CD33 and TREM2 and in the combination of the two is one of our goals. We are generating iPSCs from patients already characterized from a clinical point of view. We will differentiate these iPSCs into pyramidal neurons, which are the neuronal populations most vulnerable to dementia, and/or microglia. These cell lines will provide an important non-invasive tool for future testing of neuroprotective drugs.
CRISPR-Cas9. We are using genome editing by CRISPR-Cas9 to generate cellular models. We are already developing Progranulin (PGRN) knock-out cells and using them in vitro to test unknown regulatory functions and pathological mechanisms caused by its deficiency. We are working on signal pathway identification associated with progranulin (PGRN) and neuronal survival to highlight the role of PGRN in the pathogenesis of Frontotemporal dementia (FTD) pathogenesis.
Rendina et al.2020
Key Pubblications
- Aliperti V, Vitale E, Aniello F, Donizetti A. LINC00473 as an Immediate Early Gene under the Control of the EGR1 Transcription Factor. Noncoding RNA. (2020) 12;6(4):46. doi: 10.3390/ncrna6040046. PMID: 33198374; PMCID: PMC7712511.
- Costa B, Manzoni C, Bernal-Quiros M, Kia DA, Aguilar M, Alvarez I, Alvarez V, Andreassen O, Anfossi M, Bagnoli S, Benussi L, Bernardi L, Binetti G, Blackburn D, Boada M, Borroni B, Bowns L, Bråthen G, Bruni AC, Chiang HH, Clarimon J, Colville S, Conidi ME, Cope TE, Cruchaga C, Cupidi C, Di Battista ME, Diehl-Schmid J, Diez-Fairen M, Dols-Icardo O, Durante E, Flisar D, Frangipane F, Galimberti D, Gallo M, Gallucci M, Ghidoni R, Graff C, Grafman JH, Grossman M, Hardy J, Hernández I, Holloway GJT, Huey ED, Illán-Gala I, Karydas A, Khoshnood B, Kramberger MG, Kristiansen M, Lewis PA, Lleó A, Madhan GK, Maletta R, Maver A, Menendez-Gonzalez M, Milan G, Miller B, Mol MO, Momeni P, Moreno-Grau S, Morris CM, Nacmias B, Nilsson C, Novelli V, Öijerstedt L, Padovani A, Pal S, Panchbhaya Y, Pastor P, Peterlin B, Piaceri I, Pickering-Brown S, Pijnenburg YAL, Puca AA, Rainero I, Rendina A, Richardson AMT, Rogaeva E, Rogelj B, Rollinson S, Rossi G, Rossmeier C, Rowe JB, Rubino E, Ruiz A, Sanchez-Valle R, Sando SB, Santillo AF, Saxon J, Scarpini E, Serpente M, Smirne N, Sorbi S, Suh E, Tagliavini F, Thompson JC, Trojanowski JQ, Van Deerlin VM, Van der Zee J, Van Broeckhoven C, van Rooij J, Van Swieten JC, Veronesi A, Vitale E, Waldö ML, Woodward C, Yokoyama J, Escott-Price V, Polke JM, Ferrari R; International FTD-Genetics Consortium (2020). C9orf72, age at onset, and ancestry help discriminate behavioral from language variants in FTLD cohorts. Neurology. 2020 Dec 15;95(24):e3288-e3302. doi: 10.1212/WNL.0000000000010914. Epub 2020 Sep 17. PMID: 32943482; PMCID: PMC7836664.
- Rendina A, Drongitis A, Donizetti A, Fucci L, Milan G, Tripodi F, Giustezza F, Postiglione A, Pappatà S, Ferrari R, Bossù P, Angiolillo A, di Costanzo A, Caiazzo M, Vitale E CD33 and SIGLECL1 Immunoglobulin Superfamily Involved in Dementia (2020). J Neuropathol Exp Neurol. ;79(8):891-901. doi:10.1093/jnen/nlaa055
- Picillo M, Vitale E, Rendina A, Donizetti A, Aliperti A, Tepedino MF, Dati G, Ginevrin M, Valente EM, Barone P (2020) Clinical and Molecular Characterization of a Novel Progranulin Deletion Associated with Different Phenotypes : JOURNAL OF ALZHEIMER’S DISEASE Volume 76, Number 1:341-347 DOI: 10.3233/JAD-200151
- Aliperti V, Sgueglia G, Aniello F, Vitale E, Fucci L, Donizetti A (2019): Identification, Characterization, and Regulatory Mechanisms of a Novel EGR1 Splicing Isoform. Int J Mol Sci. Mar 28;20(7). pii: E1548. doi: 10.3390/ijms20071548.
- Italiani, P., Puxeddu, l., Napoletano, S., Scala, E., Melillo, D., Manocchio, S., Angiolillo, A., Migliorini, P., Boraschi, D., Vitale, E., Di Costanzo, A. (2018): Circulating levels of IL-1 family cytokines and receptors in Alzheimer’s disease: new markers of disease progression? J Neuroinflammation. 2018 Dec 12;15(1):342. doi: 10.1186/s12974-018-1376-1.
- Milan, G., Napoletano, S., Pappatà, S., Gentile, M.T., Colucci-D’Amato, L.,Della Rocca, G., Maciag, A., Rossetti, C.P., Fucci, L., Puca, A., Grossi, D., Postiglione, A., Vitale, E. (2017): GRN deletion in Familial Fronto-Temporal Dementia showing Association with clinical Variability in three familial cases, Neurobiology of Aging doi: 10.1016/j.neurobiolaging.2016.12.030.
- Paris, D.,Melck, D., Longo, A., Napoletano, S., Carotenuto, G., Nicolais, L., Motta, A., Vitale, E. (2016): Metabolic response of SH-SY5Y cells to gold nanoparticles by NMR-based metabolomics analyses. Biomedical Physics and Engineering Express (BPEX)
- Imperatore, R., Carotenuto, G., Di Grazia, M.A., Ferrandino, I., Palomba, L., Mariotti, R., Vitale, E., De Nicola, S., Longo, A., Cristino, L. L. (2015): Imidazole-stabilized gold nanoparticles induce neuronal apoptosis: An in vitro and in vivo study. J Biomed Mater Res A. 103:4. doi: 10.1002/jbm.a.35289.
- Polverino, A., Longo, A., Donizetti, A., Drongitis, D., Frucci, M., Schiavo, L.,Carotenuto, G., Nicolais, L., Piscopo, M., Vitale, E., Fucci , L. (2014): Molecular responses of cells to 2-mercapto-1-methylimidazole gold nanoparticles (AuNPs)-mmi: investigations of histone methylation changes. Journal of Nanoparticle Research 16:7 Article Number: 2516 DOI: 10.1007/s11051-014-2516-x
